A gentle introduction to persistent homology
In the context of our recent publications at ICLR 2019 (Neural Persistence) and ICML 2019 (Persistent Weisfeiler-Lehman), I want to shed some light on the intuition behind peristent homology, a method from Topological Data Analysis (TDA) we made use of. Thanks to Bastian Rieck for the visualizations for figure 1, and for introducing me to TDA!
Persistent Homology
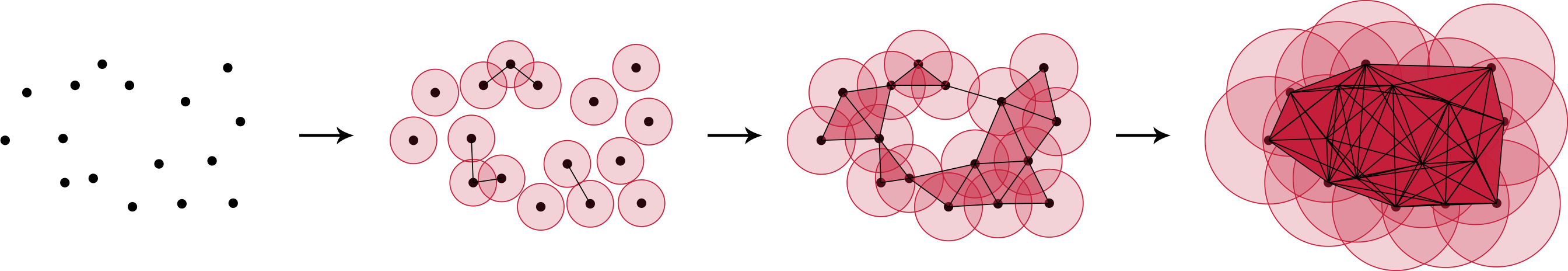
The central object in algebraic topology is a simplicial complex $K$, e.g. an undirected and weighted connected graph. Persistent homology captures topological features of $K$ on different scales by defining a filtration on it. A filtration is a nested sequence of simplicial complexes (e.g. subgraphs $K_m$) representing the “growth” of $K$: $\emptyset=K_0 \subseteq K_1 \subseteq \dots \subseteq K_{m-1} \subseteq K_m = K$. Figure 1 illustrates this growth process for a $2$-dimensional point cloud. With a defined metric in our space (e.g. the Euclidean distance), we keep track of the generation and destruction of connected components (CC) while increasing the distance threshold (i.e. a point’s range of vision). We connect two points once their gazes meet (how romantic) and hence create a CC. Note, that in this example we consider each point in the $0^{th}$ filtration-step (left most panel in figure 1) as its own connected component. Increasing the threshold therefore leads to a decreasing number of connected components. To be more precise, we start with 16 CCs, observe 11 CCs at step 1, and arrive at 1 CC at step 2 and 3.
The Persistence Diagram
Above, I mentioned that we “keep track” of the creation and destruction of CCs during the filtration. Figure 2 illustrates this on a graph $G=(V,E)$ with a slightly adjusted filtration setup: Earlier, all points in our space represented their own connected component at step $m=0$. Now, a node $n$ is only created at threshold $\epsilon_m$ when an edge $e$ of the form $(v, n)$ or $(n, v)$, with $v \in V \setminus \{n\}$ exists and $\text{weight}(e) \leq \epsilon_m$. In other words, we initialize each node with the minimum value of its edges and use the edge weights (rather than radii) to determine whether a CC is created.
We keep track of the birth (creation) $\epsilon_{birth}$ and death (destruction) $\epsilon_{death}$ thresholds of all CCs and put the tuples ($\epsilon_{birth}, \epsilon_{death}$) in a diagram – the persistence diagram $\mathcal{D}$ – where the $x$-Axis represents birth and the $y$-Axis death (how dark). Let’s take a closer look at figure 2 to understand how a persistence diagram comes into existence:
- The first two CCs (orange and green) are generated at threshold 1. Hence, the first two tuples to be added to the PD are of the form $(1, \tilde{\epsilon}_{death}=1)$. $\tilde{\epsilon}_{death}$ is a placeholder for visualization purposes only, that allows us to update the PD at each threshold. It always equals the current threshold.
- At threshold $2$ another CC is generated. We populate the PD with $(2, \tilde{\epsilon}_{death}=2)$
- The first interesting event happens at threshold $3$. The green CC becomes part of the orange one. This is when green’s time has come and we update the respective tuple to $(1,\epsilon_{death}=3)$. The survivor lives on and gets ready to swallow red.
- This happens at threshold $4$. We updated red’s tuple to $(2,\epsilon_{death}=4)$ and see how it continues its triumph up to the largest threshold.
- The diagram at threshold $5$ is the PD of graph $G$.
Given a point $(x,y)$ from the persistence diagram $\mathcal{D}$, the persistence of the connected component which is represented by this tuple is defined as $$\text{pers}(x,y) := |y-x|.$$ A central observation that was made by Edelsbrunner et al. in Topological persistence and simplification, is that high persistence is considered to correspond to features, while low persistence is considered to indicate noise. Furthermore, we can summarize these topological features of a persistence diagram by its $p$-norm: $$ ||\mathcal{D}||_p:= \Big(\sum_{(c,d)\in \mathcal{D}}pers(c,d)^p\Big)^{\frac{1}{p}} $$
I hope this post helped you to get an idea of what persistent homology is and you might already ponder about how to apply this to deep learning. Feel free to check out our paper to learn how to derive an early stopping criterion that does not rely on validation data using persistent homology.